| Equipe 10 avenir : Michele Trabucchi | |||
| CONTROL OF GENE EXPRESSION | |||
| Previous activities | |||
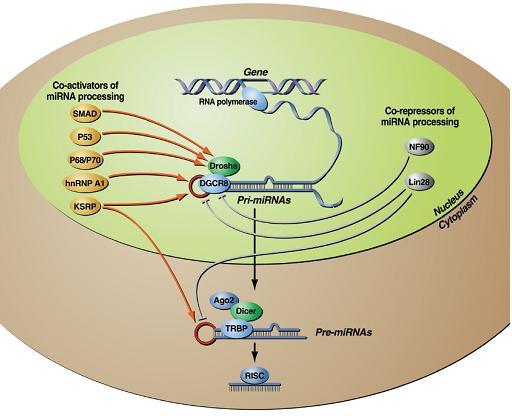
Our research is focused on the molecular mechanisms underlying gene expression control at the post-transcriptional level in response to extracellular signals. Two post-transcriptional pathways mainly control the gene expression, namely microRNAs (miRNAs) and AU-rich element Mediated Degradation of mRNA (AMD). MicroRNAs (miRNAs) are evolutionary conserved small non-coding RNAs, which arise from processing of hairpin primary transcripts by two sequential RNAse III domain-containing enzymes, namely Drosha and Dicer, to generate a mature form of about 22 nucleotides. Mature miRNAs are loaded in the RNA-induced silencing complex (RISC), containing Argonaute 2, in order to mediate degradation and/or inhibition of translation of specific target mRNAs via Watson-Crick base pairing partial sequence complementary. Expression of many miRNAs is consistently tissue or developmental stage specific, and its deregulation is involved in numerous human diseases, including genetic disorders, such as chronic lymphocytic leukaemia. Besides miRNAs, AMD is another pathway aiming to post-trascriptionally control the gene expression. AMD promotes decay of unstable mRNAs which possess crucial biological functions, e.g. cyclins and B-catenin mRNAs. AMD requires at least three components, (i) an instability element, such as the Adenylate/uridylate−Rich Elements (ARE) located in the 3’ untranslated region (3’ UTR), (ii) ARE binding proteins (ARE−BPs), and (iii) two enzymes, the exosome and the deadenylation enzyme PARN. ARE−BPs, such as AUF1, TTP, TIA-1, KSRP (KH-type splicing regulatory protein) promote decay of ARE containing RNAs, while HuR stabilizes them. Importantly, both miRNA and AMD pathways represent a very established cellular strategy that permits relatively quick changes in gene expression pattern in response to environmental or developmental stimuli.
Different reports pointed out the relevance of both transcriptional and post-transcriptional regulation of miRNAs expression demonstrating a plethora of regulatory options to differentially control miRNA levels. We have published a paper in Nature describing a novel mechanism of microRNA processing regulation. In particular, we showed that KSRP is an integral component of both Drosha and Dicer complexes and promotes the maturation of a subset of miRNA precursors. Our data indicate that activators of miRNA processing (KSRP and possibly additional RNA binding proteins) as well as repressors of miRNA processing (e.g. Lin28 for let-7 precursors), act in a coordinated way to convert cellular signals into changes in miRNA expression processing, providing the first rational basis to identify additional repressors and activators of miRNA biogenesis. Among miRNAs whose maturation is controlled by KSRP, we have found miR-155 (published in FASEB J), a microRNA that is up-regulated in immune cells upon microbial and inflammatory challenges. miR-155 regulates the magnitude of the innate inflammatory response by modulating different inflammatory transcripts. We found that LPS treatment regulates the maturation of miR-155 leaving unaffected pri-miR-155 transcription, indicating the existence of a LPS-induced and KSRP-dependent post-transcriptional regulation of miR-155 biogenesis. Together, these data infer a continuum of KSRP-dependent molecular strategies for RNA regulation: promoting in one hand the biogenesis of microRNAs and in the other hand the degradation of unstable mRNAs, in each case based on its versatile ability to recognize different classes of RNA motifs.
|
|||
| Research Projects | |||
Uncontrolled and chronic inflammation is nowadays considered a major component of many widely occurring diseases, including asthma, atherosclerosis and peripheral vascular disease, Alzheimer’s disease, type 2 diabetes, and cancer. Innate immune and inflammatory response significantly contributes to onset, progression, tissue dysfunction, and ultimately organ failure in these diseases. The study of the mechanisms regulating the amplitude and the consecutive resolution of the inflammation may provide new insight into the pathogenesis of inflammatory diseases allowing the exploration of novel targets for the treatment and the prevention of many disorders. | |||
| Major Publications | |||
|
*equally contribution 2015 Saccani S, Trabucchi M. Regulation of stimulus-inducible gene expression in myeloid cells. Semin Immunol. 2015 Feb;27(1):33-43 2012 E. Repetto, P. Briata, N. Kuziner, B.D. Harfe, M.T. McManus, R. Gherzi, M.G. Rosenfeld, M. Trabucchi (2012) Let-7b/c enhance the stability of a tissue-specific mRNA during mammalian organogenesis as part of a feedback loop involving KSRP. Plos Genetics Jul;8(7): e1002823. Q. Hu, B. Tanasa*, M. Trabucchi*, W. Li, J. Zhang, K.A. Ohgi, D.W. Rose, C.K. Glass, M.G. Rosenfeld (2012) DICER- and AGO3-dependent generation of retinoic acid-induced DR2 Alu RNAs regulates human stem cell proliferation. Nature structural & molecular biology 19, 1168-1175
2011 P. Briata, W.J. Lin, M. Giovarelli, M. Pasero, C.F. Chou, M. Trabucchi, M.G. Rosenfeld, C.Y. Chen, R. Gherzi (2011). PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis. Cell Death & Differ Sep 2. doi: 10.1038/cdd.2011.117. [Epub ahead of print] P. Briata, C.Y. Chen, M. Giovarelli, M. Pasero, M. Trabucchi, A. Ramos, R. Gherzi (2011). KSRP, many functions for a single protein. Front Biosci. 2011 Jan 1;16:1787-96 2010 R. Gherzi*, M. Trabucchi*, M. Ponassi, I.E. Gallouzi, M.G. Rosenfeld, P. Briata (2010). Pitx2, a required factor in myoblast differentiation, is phosphorylated by AKT and controls cyclin D1 mRNA decay. Cell Death & Differ 17(6):975-983. 2009 M. Trabucchi, P. Briata, M.F. Garcia-Mayoral, A.D. Haase, W. Filipowicz, A. Ramos, R. Gherzi, M.G. Rosenfeld (2009). The RNA-binding Protein KSRP Promotes the Biogenesis of a Subset of miRNAs. Nature 459:1010-1014. T. Ruggiero*, M. Trabucchi*, F. De Santa, S. Zupo, B.D. Harfe, M.T. McManus, M.G. Rosenfeld, P. Briata, R. Gherzi (2009). LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J 23:2898-2908. M. Trabucchi*, P. Briata*, W. Filipowicz, M.G. Rosenfeld, A. Ramos, R. Gherzi (2009). How to control miRNA maturation? Co-activators and co-repressors take the stage. RNA Biology Nov-Dec;6(5):536-40 (Review) 2008 M. Trabucchi, V.L. Trudeau, H. Tostivint, I. Lihrmann, M. Vallarino, H. Vaudry (2008). Molecular characterization and comparative localization of the mRNAs encoding two glutamic acid decarboxylases (GAD65 and GAD67) in the brain of the African lungfish, Protopterus annectens. J Comp Neurol 506:979-988 2007 T. Ruggiero*, M. Trabucchi*, M. Ponassi, G. Corte, C.Y. Chen, L. al-Haj, K.S. Khabar, P. Briata, R. Gherzi (2007). Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling. BMC Mol Biol. Apr 16;8:28. 2006 R. Gherzi, M. Trabucchi, M. Ponassi, T. Ruggiero, G. Corte, C. Moroni, C.Y. Chen, K.S. Khabar, J.S. Andersen, P. Briata (2006). The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 5:e5. |
|||
 |
|||
| Home | Top of page | Publications | |
| Team Leader | ||||||
|---|---|---|---|---|---|---|
 |
||||||
| mtrabucchi@unice.fr | ||||||
| Researcher | ||||||
| Postdoc | ||||||
| Student | ||||||
| Research assistant | ||||||
|
||||||